
YAPiC is developed by Image and Data Analysis Facility, Core Reseach Facilities of DZNE(German Center for Neurodegenerative Diseases).
-
Getting Started I: Train Your First Model
-
Getting Started II: Apply your model in Fiji
-
Documentation
What is YAPiC for?
With YAPiC you can make your own customized filter (also called model or classifier) to enhance a certain structure of your choice with a simple python based command line interface, installable with pip:
$ yapic train unet_2d "path/to/my/images/*.tif" path/to/my/labels.ilp
$ yapic predict my_trained_model.h5 path/to/results/
You can, e.g., train a model for detection of oak leaves in color images, and use this oak leaf model to filter out all image regions that are not covered by oak leaves:
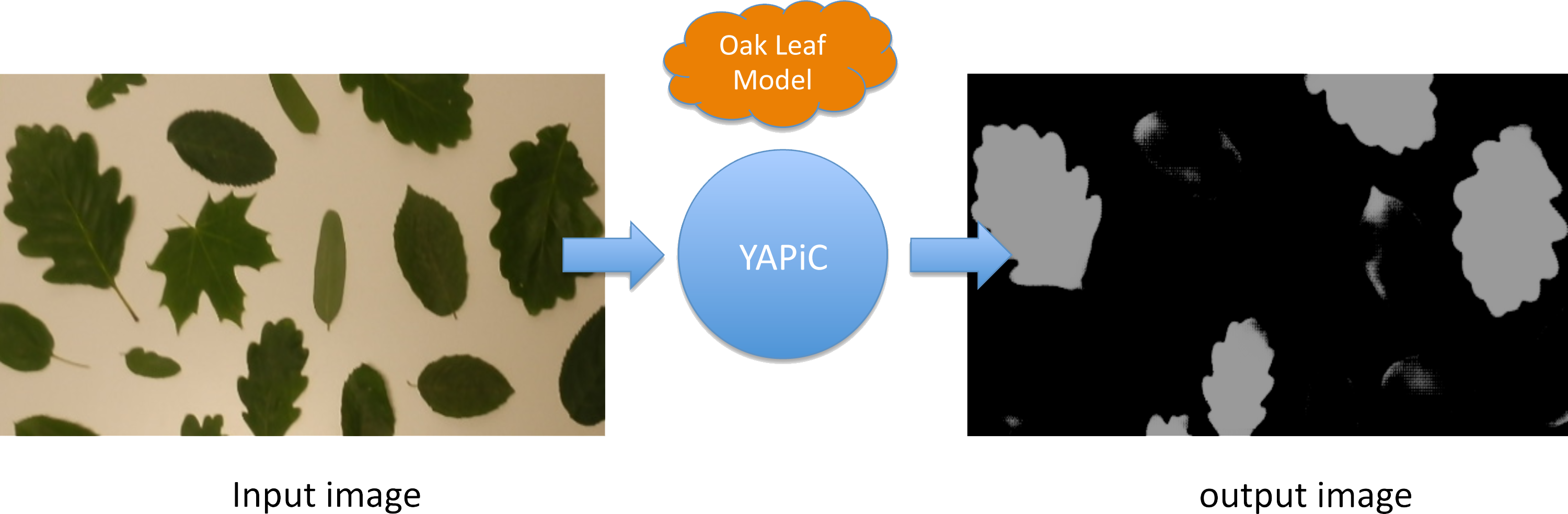
- Pixels that belong to other leaf types or to no leaves at all are mostly suppressed, they appear dark in the output image.
- Pixels that belong to oak leaves are enhanced, they appear bright in the output image.
The output image is also called a probability map, because the intensity of each pixel corresponds to the probability of the pixel belonging to an oak leaf region.
You can train a model for almost any structure you are interested in, for example to detect a certain cell type in histological micrographs (here: purkinje cells of the human brain):
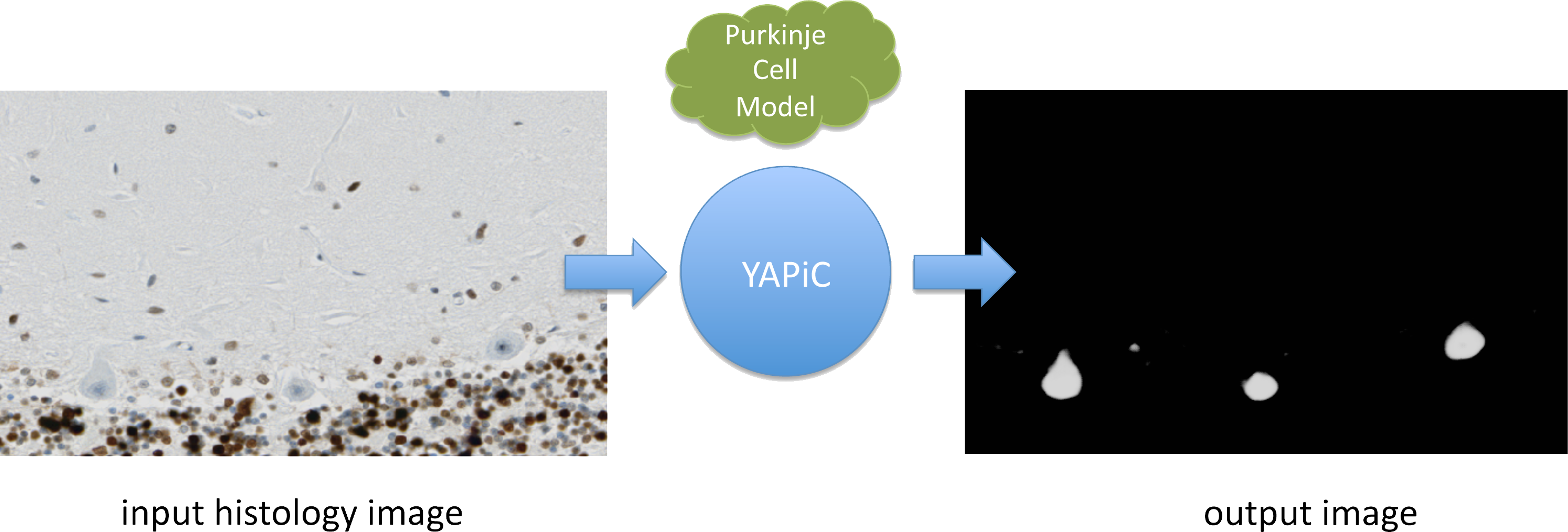 Histology data provided by Oliver Kaut (University Clinic Bonn, Dept. of Neurology)
Histology data provided by Oliver Kaut (University Clinic Bonn, Dept. of Neurology)
We have used YAPiC for analyzing various microscopy image data. Our experiments are mainly related to neurobiology, cell biology, histopathology and drug discovery (high content screening). However, YAPiC is a very generally applicable tool and can be applied to very different domains. It could be used for detecting, e.g., forest regions in satellite images, clouds in landscape photographs or fried eggs in food photography.
Examples
- Live cell imaging: Detection of neurites in label-free time lapse imaging.
- Digital Pathology: Detection of specific cell types in histological micrographs.
- Electron Microscopy: Detection of actin filaments in transmission electron micrographs.
Why YAPiC?
Pixel classification in YAPiC is based on deep learning with fully convolutional neural networks. Development of YAPiC started in 2015, when Ronneberger et al. presented a U-shaped fully convolutional neural network that was capable of solving highly challenging pixel classification tasks in bio images, such as tumor classification in histological slides or cell segmentation in brightfield DIC images.
YAPiC was designed to make this new kind of AI powered pixel classification simply applicable, i.e feasible to use for a PhD student in his/her imaging project.
Simply applicable means here in detail:
- Easy to install.
- Working out of the box with 3D multichannel images saved with Fiji.
- Easy collection of label data by utilizing the great Ilastik user interface.
- Support of sparse labels. From our experience, labels can be collected within a few hours by one single person.
- Simple command line and programming interface (Python).
How to install
Linux
-
Install Python 3.6.
- Update pip and setuptools
pip install --upgrade pip setuptools -
Install Tensorflow
- We strongly recommend to install a Tensorflow version with GPU support. CPU is too slow for model training.
- Please read Tensorflow installation instructions to set up CUDA drivers, cuDNN etc. correctly.
- The support for Tensorflow 1 is discontinued. We only support Tensorflow 2.
- To be able to export YAPiC models to ImageJ (DeepImageJ Plugin version 1.2), you have to install
Tensorflow version 1.15.
pip install tensorflow-gpu==1.15Hint: You can make different virtual environments with different Tensorflow versions for model training and ImageJ export.
- You can train your model with an environment where
tensorflow==2.1is installed. If you have a very new CUDA driver, this may increase training speed compared to older Tensorflow versions. - For exporting the model to ImageJ, you can switch to an environment with
tensorflow==1.15(GPU support is not necessary for just exporting the model).
- You can train your model with an environment where
- Install YAPiC
We recommend to install legacy YAPiC version which supports Ilastik label files. The current YAPiC version does not support Ilastik any more. Instead a label and prediction workflow with napari is supported. However the napari workflow for labelling, YAPiC model training and prediction has not been documented so far and is not working robustly.
The tutorials you find here in the documentations work only with yapic_io<=0.1.2 and yapic<=1.2.3.
pip install tensorflow==2.4.1 # you may use other tensorflow versions regarding to your hardware
pip install yapic_io==0.1.2 # last version supporting Ilastik label files and producing reliable output
pip install yapic==1.2.3 # newer yapic versions require yapic_io>0.2.3
Windows and Mac
YAPiC is currently only supported on Linux. It runs, in principle, on Mac OS, but installing Tensorflow with GPU support in currently not that straightforward on Mac OS. We may release Docker images in the future to run YAPiC easily in Windows and Mac workstations.
On Windows you may use the Windows Subsystem for Linux and install CUDA drivers, Tensoerflow and YAPiC on a Ubuntu subsystem.
How to use it
Command line interface
Model training
yapic train unet_2d "path/to/my/images/*.tif" path/to/my/labels.ilp
Prediction
yapic predict my_trained_model.h5 path/to/results/
- Get hands on YAPiC with the tutorial.
- Go to CLI Documentation for more details.
Apply your trained model in Imagej/Fiji
Once you have trained a model, you can convert the model with YAPiC to use it with the DeepImageJ plugin of ImageJ/Fiji.
yapic deploy my_trained_model.h5 path/to/example/image.tif path/to/my/deepimagej_model
For deployment to DeepimageJ you have to use Tensorflow version 1.13.1 (see installation instructions below)
- Get hands on YAPiC with the tutorial.
- Go to CLI Documentation for more details.
Python API
Model training
from yapic.session import Session
img_path = 'path/to/my/images/*.tif'
label_path = 'path/to/my/labels.ilp'
model_size_zxy = (5, 572, 572)
t = Session()
t.load_training_data(img_path, label_path)
t.make_model('unet_multi_z', model_size_zxy)
t.define_validation_data(0.2) # 80% training data, 20% validation data
t.train(max_epochs=5000,
steps_per_epoch=48,
log_filename='log.csv')
t.model.save('my_model.h5')
Prediction
from yapic.session import Session
img_path = 'path/to/my/images/*.tif'
results_path = 'path/to/my/results/'
t = Session()
t.load_prediction_data(img_path, results_path)
t.load_model('my_model.h5')
t.predict() # applies the classfier to all images in img_path
Try it out with the leaves example dataset
Hardware Recommendations
YAPiC is designed to run on dedicated hardware. In production, it should not run on your everyday-work notebook, but on a dedicated workstation or a server. The reason is, that training a model requires long time (multiple hours to multiple days) and a lot of computing power. Running these processes in the background on your notebook while e.g. writing E-Mails is not feasible. Moreover, you will need powerful GPU hardware that is normally not available on office notebooks.
- Using fast SSD hard drives (PCIe SSDs) for storing training data may increase training speed, compared to conventional hard drives. Have a look at the GPU requirements for Tensorflow.
- From our expericence you can have already quite good performance with NVIDIA Geforce boards (mainly intended for gaming). These are cheaper than professional NVIDIA Tesla GPUs.
- GPU RAM requirements: RAM of your GPU hardware is often a bottleneck and depends the specific project. RAM requirements depend on the number of classes you want to train and if you use a 2D network or 3D network. Some recommendations, based on our personal experience:
- For training a unet_2D with two classes (foreground, background), 5 GB RAM on your GPU is sufficient.
- For training a unet_multi_z with five z-layers and two classes, 11 GB RAM on GPU is sufficient.