Detection of purkinje cells in histological brain slices
This project was executed by Christoph Moehl (Core Research Facilities, DZNE Bonn) and Oliver Kaut (University Clinic Bonn, Dept. of Neurology).
Objective and outcome
In the following example we aimed to locate and quantify
- individual purkinje cells
- granule cell layer regions
- cells of connective tissue
in human brain slices.
By using unet_2d we trained a classifier in YAPiC with 4 classes:
- purkinje cells (red)
- granule cell region (green)
- connective tissue cells (blue)
- background (black)
Look at the image below: On the upper part, you see the raw input image. Below you see the YAPiC output: The four purkinje cells are clearly enhanced (red signal).
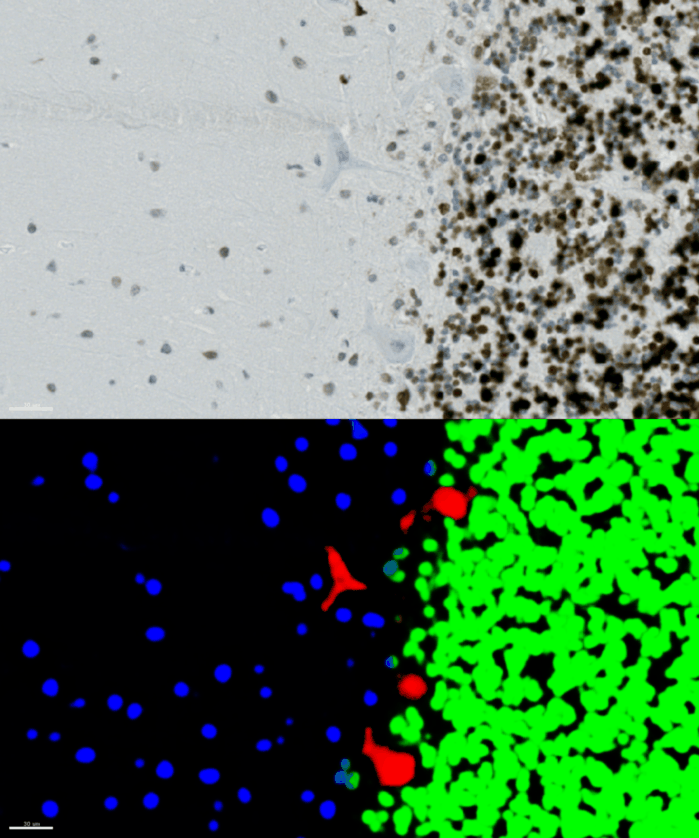
Histology data by Dr. Oliver Kaut, University Clinic Bonn, Dept. of Neurology
With YAPiC you can process huge histological datasets in bigtiff format. The image above is just a small subset (the yellow box) of the whole image:
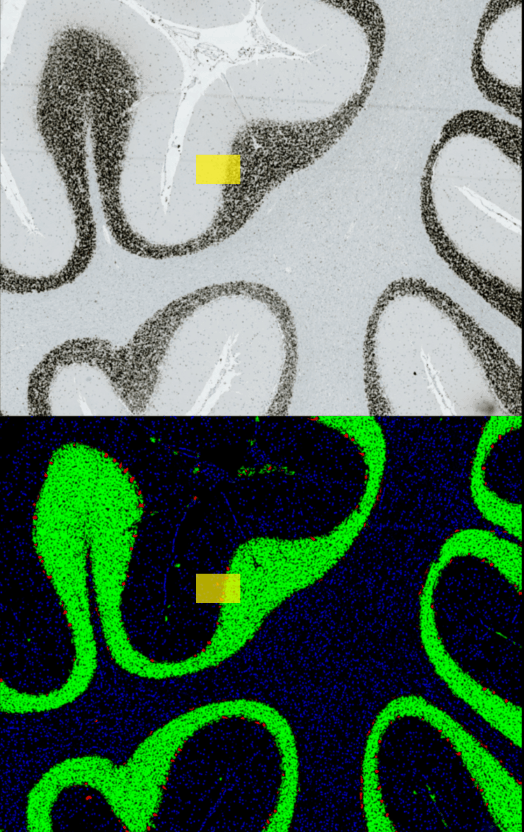
Histology data by Dr. Oliver Kaut, University Clinic Bonn, Dept. of Neurology
How to proceed from here?
You could count and measure individual purkinje cell objects by using identifyPrimaryObjects module of CellProfiler.
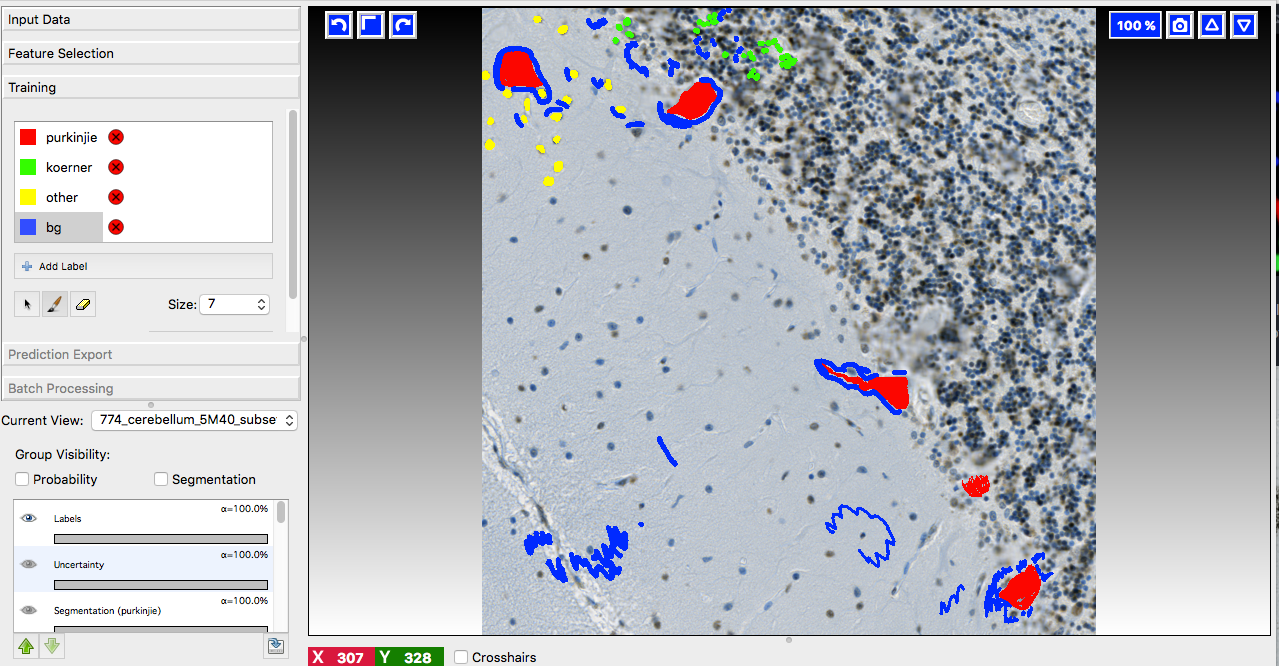
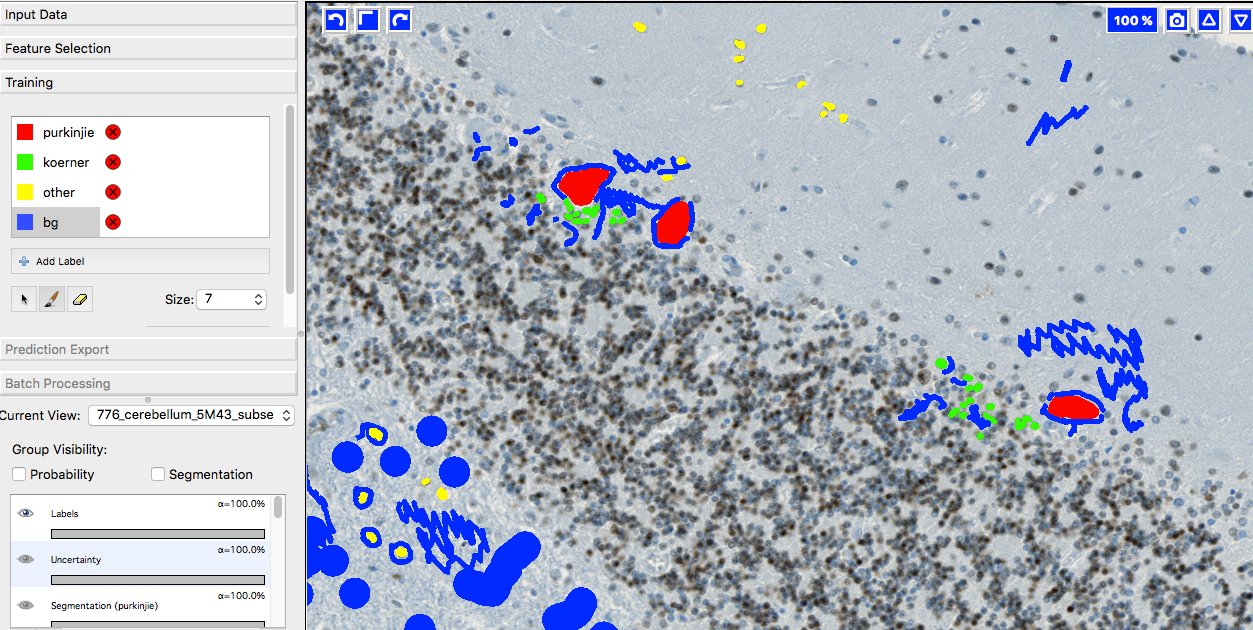
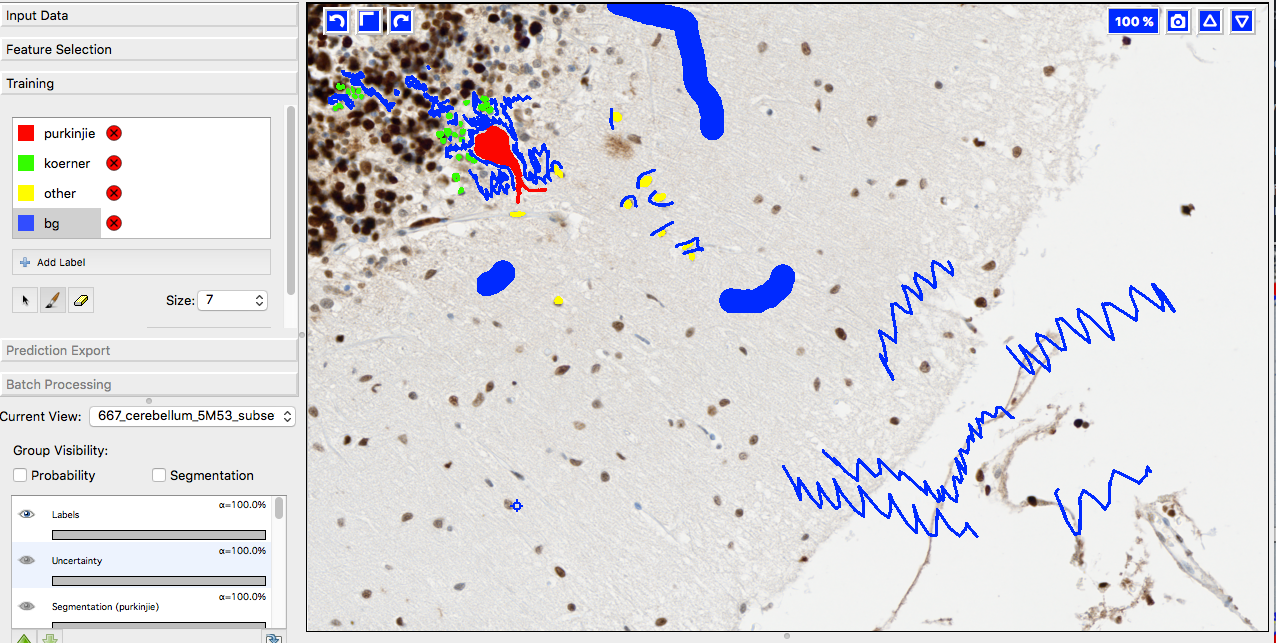
A few words on training data collection
We prepared 36 image subsets from ca 20 different tissue slide scans and imported them into ilastik for labeling. YAPiC reads label data directly from ilastik project files (ilp).
Below you see 4 examples from the 36 labeled training images. Note the following about the labeling strategy:
- We did not label whole image. We rather labeled many images sparsely than a few images very densely.
- Borders between classes are clearly defined.
- Amounts of labels are more or less balanced between all classes.